Congenital heart disease (CHD) is the most common type of congenital malformation, arising from structural or functional abnormalities during early embryonic heart development. A deep understanding of the underlying molecular mechanisms and developmental processes is crucial for improving the diagnosis, treatment, and prevention strategies for CHD. Heart development relies on precise spatiotemporal regulatory mechanisms; however, many mysteries still surround the spatial origin and spatiotemporal dynamic regulation of cardiac progenitor cells.
On June 10, 2025, the team of Professor Lin Chengqi and Professor Luo Zhuojuan from the School of Life Science and Technology, and Research Professor Xie Peng from the School of Biological Science and Medical Engineering, in collaboration with several research institutions, published a research paper in the journal Genome Biology titled, "Integration of single-cell and spatial transcriptomics by SEU-TCA reveals the spatial origin of early cardiac progenitors." The paper introduces a novel integrative analysis tool for spatial transcriptomics—SEU-TCA (Spatial ExpressionUtility - Transfer Component Analysis)—which deciphers the "spatial information code" of cardiac progenitors and systematically reveals spatial subtypes of second heart field progenitor cells.
The fate determination of cardiac progenitor cells is closely linked to their precise spatial localization. However, traditional methods cannot achieve spatial information analysis with single-cell precision, making it difficult to accurately define progenitor cell subtypes and their spatial distribution. To overcome this challenge, the research team proposed an integration strategy based on Transfer Component Analysis (TCA), named SEU-TCA. By jointly analyzing single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) data, this method achieves accurate spatial mapping of single cells. The team systematically evaluated the performance of SEU-TCA and compared it with other algorithms using various biological samples from different technology platforms (mouse gastrula, human heart, mouse olfactory bulb, human pancreatic ductal adenocarcinoma), demonstrating SEU-TCA's superior capabilities in spatial deconvolution and single-cell localization. Particularly in the integration of scRNA-seq data and spatial atlases of mouse E7.5 embryos, SEU-TCA successfully established a correlation between gene expression levels and spatial positions, thereby revealing multiple spatially specific mesodermal cell subtypes.
Through a combined analysis using SEU-TCA and regulon activity, the team systematically unveiled spatial subtypes of second heart field (SHF) cardiac progenitors. They identified the specific expression and regulatory activity of the Irx family oftranscription factors in the c2 subpopulation of anterior second heart field (aSHF) progenitors, proposing that IRX1 may be a key regulatory factor for this subpopulation. Through genetic lineage tracing, it was confirmed that IRX1-positive cells contribute to the construction of the outflow tract (OFT) and the right ventricle (RV). Furthermore, using conditional knockout technology, they found that the deletion of Irx1 in cardiac progenitor cells leads to ventricular septal defects (VSDs). This discovery provides a new theoretical basis for understanding the molecular foundations of congenital heart disease.
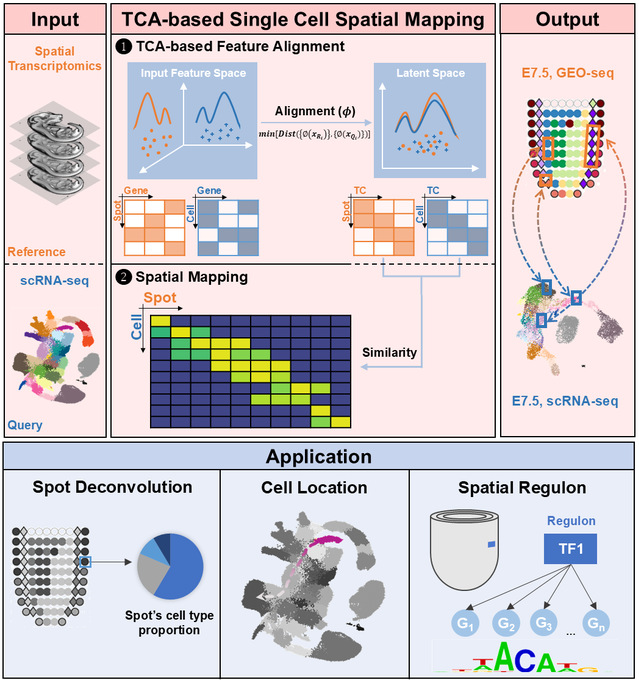
This study provides a new, efficient method for integrating spatial omics and single-cell omics data. It is the first to reveal a cardiac progenitor subtype regulated by IRX1, offering a new target for the diagnosis of congenital heart disease. The broad applicability and excellent predictive accuracy of SEU-TCA will aid in-depth research in fields such as organ development, tissue regeneration, and tumor spatial heterogeneity.
The co-first authors of the paper are He Jingjing, a doctoral student at the School of Life Science and Technology, Southeast University; Dr. Yang Yi, an Associate Professor at the same school; Jiang Rui, a doctoral student at Tongji University; and Zheng Yanying, a doctoral student at the School of Life Science and Technology, Southeast University. The co-corresponding authors are Professor Lin Chengqi from the School of Life Science and Technology, Southeast University; Research Professor Xie Peng from the School of Biological Science and Medical Engineering; Professor Wei Ke from Tongji University; Professor Luo Zhuojuan from the School of Life Science and Technology, Southeast University; and Chief Physician Cao Hailong from Zhongda Hospital, Southeast University. Professor Yang Zhongzhou from Nanjing University, Professor Jing Naihe and Associate Research Professor Yang Xianfa from Guangzhou Laboratory also participated in this research. This work was supported by the National Key R&D Program of China.
Paper Link:https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03633-3
